Answer: a bivariate dataset is a dataset that contains two variables.
Answer: a bivariate normal dataset is a bivariate dataset that is normal in every direction. In particular, if x and y are the two variables in the dataset, then both x and y are normally distributed. A parsimonious description of a bivariate normal dataset requires five statistics: X, SD+x, Y, SD+y, and the correlation r between x and y.
setwd("c:/workspace")
getwd( )
[1] "c:/workspace"
# Create a dataframe df to hold the 2026 Bears Roster
# First download the bears-2026-roster into
# the c:/workspace directory (folder).
df <- read.csv("bears-2026-roster.txt")
# Create data vector of players heights in meters:
h <- (df$HtFt + df$HtIn / 12) * 0.3048
# Create data vector of players weights in kilos:
w <- df$Weight * 0.4536
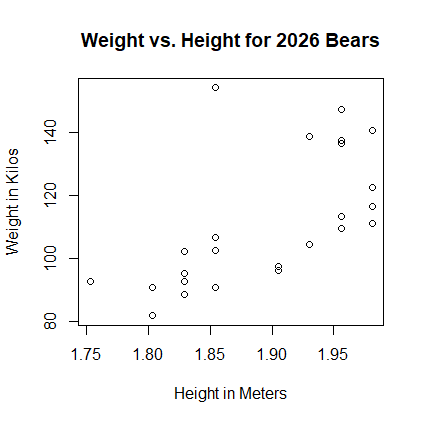
plot(h, w, xlab="Player Height (Meters)",
ylab="Player Weight (Kilos),
main="Height and Weight of 2026 Bears Roster")